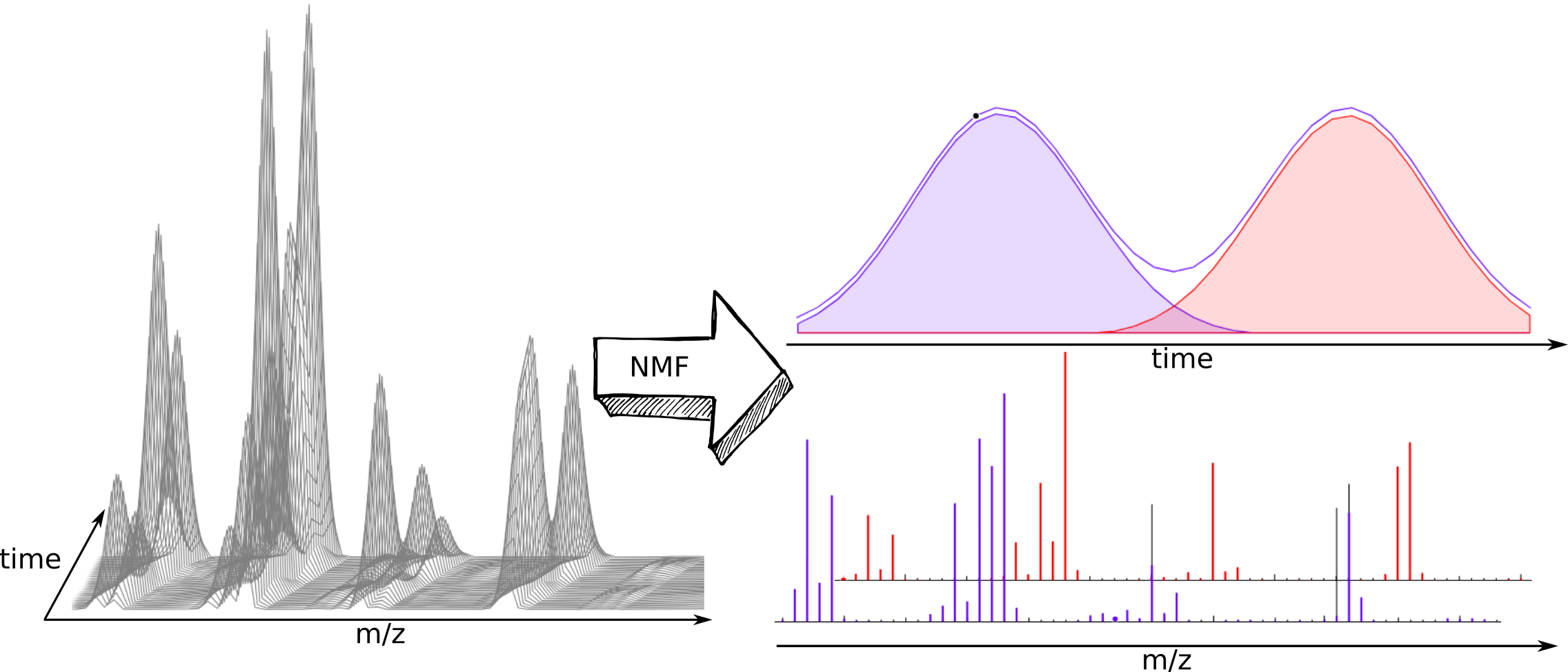
nGMCA - non-negative Generalized Morphological Component Analysis
A tool for non-negative matrix factorization.
Instalation
$ npm install ml-ngmca
Usage
import { nGMCA } from 'ml-ngmca';
const result = nGMCA(dataMatrix, options);As a CommonJS module
const { nGMCA } = require('ml-ngmca');
const result = nGMCA(dataMatrix, options);API Documentation
This algorithm is based on the article Jérémy Rapin, Jérôme Bobin, Anthony Larue, Jean-Luc Starck. Sparse and Non-negative BSS for Noisy Data, IEEE Transactions on Signal Processing, 2013.IEEE Transactions on Signal Processing, vol. 61, issue 22, p. 5620-5632, 2013.
In order to get a general idea of the problem you could also check the Wikipedia article.
Examples
You will be able to separate the components of a mixture if you have a series of measurements correlated by a composition profile e.g NMR or mass spectra coming from a chromatographic coupled technique of two or more close retention times. So you will have a matrix with a number of rows equal or greater than the number of pure components of the mixture.
import { Matrix } from 'ml-matrix';
import { nGMCA } from 'ml-ngmca';
let pureSpectra = new Matrix([[1, 0, 1, 0]]);
let composition = new Matrix([[1, 2, 3, 2, 1]]);
// matrix = composition.transpose().mmul(pureSpectra)
let matrix = new Matrix([
[1, 0, 1, 0],
[2, 0, 2, 0],
[3, 0, 3, 0],
[2, 0, 2, 0],
[1, 0, 1, 0],
]);
const options = {
maximumIteration: 200,
phaseRatio: 0.4,
};
const result = nGMCA(matrix, 1, options);
const { A, S } = result;
console.log(`A = ${A.to2DArray()} S =${S.to2DArray()}`);
/**
A = [
[ 0.22941573387056177 ],
[ 0.45883146774112354 ],
[ 0.6882472016116853 ],
[ 0.45883146774112354 ],
[ 0.22941573387056177 ]
]
S = [ [ 4.358898943540674, 0, 4.358898943540674, 0 ] ]
if you reescale both S maxS and A with 1/maxS.
*/
let maxByRow = [];
for (let i = 0; i < S.rows; i++) {
maxByRow.push(S.maxRow(i));
}
S.scale('row', { scale: maxByRow });
A.scale('column', {
scale: maxByRow.map((e) => 1 / e),
});
/**
S = [ [ 1, 0, 1, 0 ] ]
A = [
[1.0000000000000002],
[2.0000000000000004],
[3.0000000000000004],
[2.0000000000000004],
[1.0000000000000002]
]
*/
const estimatedMatrix = A.mmul(S);
const diff = Matrix.sub(matrix, estimatedMatrix);Here is a second example:
let matrix = new Matrix([
[0, 0, 1, 1, 1],
[0, 0, 1, 1, 1],
[2, 2, 2, 0, 0],
[2, 2, 2, 0, 0],
]);
const options = {
maximumIteration: 200,
phaseRatio: 0.4,
};
const result = nGMCA(matrix, 1, options);
const { A, S } = result;
console.log(`A = ${A} S =${S}`);
/**
A = [
[
0.707107 0
0.707107 0
2.26e-17 0.707107
2.26e-17 0.707107
]
]
S = [
[
9.86e-32 9.86e-32 1.41421 1.41421 1.41421
2.82843 2.82843 2.82843 0 0
]
]
note: 9.86e-32 and 2.26e-17 is practically zero
so if you reescale both S maxS and A with 1/maxS.
*/
let maxByRow = [];
for (let i = 0; i < S.rows; i++) {
maxByRow.push(S.maxRow(i));
}
S.scale('row', { scale: maxByRow });
A.scale('column', {
scale: maxByRow.map((e) => 1 / e),
});
console.log(`A = ${A} S =${S}`);
/**
A = [
[
1 0
1 0
0 1
0 1
]
]
S = [
[
0 0 1 1 1
2 2 2 0 0
]
]
*/The result has the matrices A and S, the estimated matrices of compositions and pureSpectra respectively. It's possible that the matrices A and S have not the same scale than pureSpectra and composition matrices because of AS has an infinity of combination to get the target matrix.