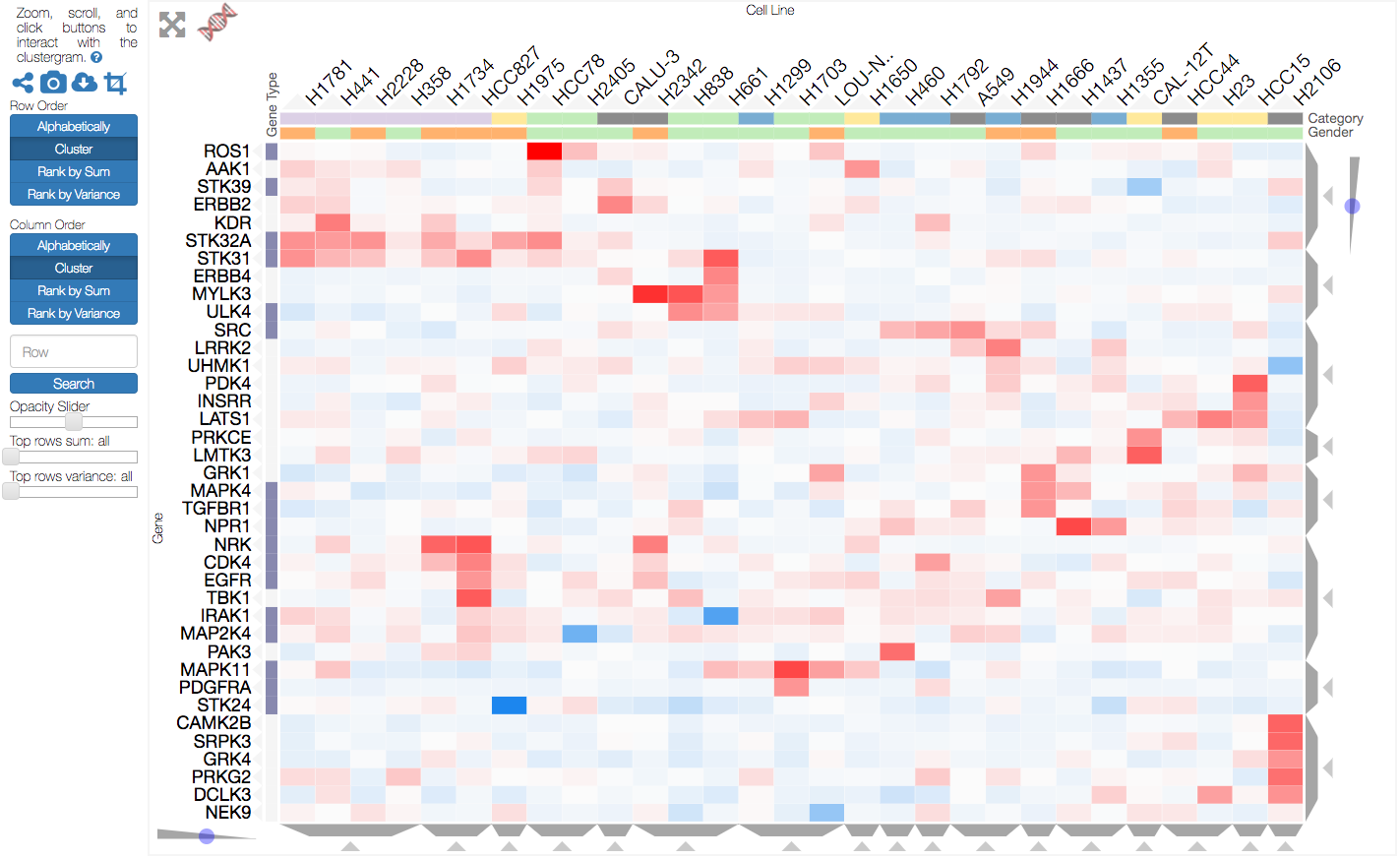
clustergrammer
Clustergrammer.js is an interactive heatmap/clustergram matrix visualization tool implemented in D3.js. The Clustergrammer web app includes an interactive demo that can be seen here (repo here) and a screenshot is show below.
The project began as an extension of this example http://bost.ocks.org/mike/miserables/ and some of clustergrammer's interacive features include:
- zooming/panning
- reordering based on sum, variance etc.
- dendrogram
- row filtering and searching
- multiple categories for rows and columns
The Clustergrammer.js source code is under the src directory and Webpack Module Developer is being used to make clustergrammer.js. The easiest way to visualize a matrix of your own data is to follow the example workflow) that uses the Clustergrammer python module (discussed here and repo here clustergrammer.py). Users can also generate the visualization json (see example mult_view.json) using another programming language as long as they adhere to the format. Clustergrammer is designed to be a reusable chart and has been integrated into several Ma'ayan lab web tools including:
clustergrammer.js
To make a visualization pass an arguments object with the following required values to Clustergrammer:
var args = {
'root':'#id_of_container',
'network_data': network_data
};
var cgm = Clustergrammer(args);
The id of the container where the visualization SVG will be placed is passed as root (this root container must be made by the user). The visualization JSON (example here mult_view.json and format discussed here) contains the information necessary to make your visualization and is passed as network_data. The visualization JSON is produced by clustergrammer.py. See additional Optional Arguments for more information.
Example Pages
The page index.html (and the corresponding script load_clustergram.js) demonstrates how to make a full-screen resizable clustergrammer visualization.
The page multiple_clust.html (and corresponding script load_multiple_clustergrams.js) demonstrates how to visualize multiple clustergrams on one page. Note that each visualization requires its own container.
Visualization Resizing
The visualization can be resized by: first resizing the container and then resizing the visualization using cgm.resize_viz(). An example of resizing when the window change size is shown below.
d3.select(window).on('resize', function(){
// first, resize the container when the screen size changes
your_function_to_resize_the_container();
// then, resize the visualization to match the container's new size
cgm.resize_viz();
});
Clustergrammer.js Dependencies
- D3.js
- jQuery
- Underscore.js
Clustergrammer Python Module
The Clustergrammer python module clutergrammer.py, takes a tab-separated matrix file as input (see format here), calculates clustering, and generates the visualization json (see format here) for clustergrammer.js. The module can be installed using pip:
pip install clustergrammer
or the source code can be obtained from clustergrammer.py repo: simply copy the clustergrammer directory with the source code to the main directory to use the module in this repo.
Example Workflow
from clustergrammer import Network
net = Network()
# load matrix file
net.load_file('txt/rc_two_cats.txt')
# calculate clustering
net.make_clust(dist_type='cos',views=['N_row_sum', 'N_row_var'])
# write visualization json to file
net.write_json_to_file('viz', 'json/mult_view.json')
The script make_clustergrammer.py is used to generate the visualization jsons for the examples pages in this repo. To visualize your own data modify the make_clustergrammer.py script.
Input Matrix Format
Clustergrammer.py discussed here takes a tab separated matrix with unique row and column names as input. The simplest format is shown here (note: that tabs are required, but spaces are used in the example below to increase readability):
Col-A Col-B Col-C
Row-A 0.0 -0.1 1.0
Row-B 3.0 0.0 8.0
Row-C 0.2 0.1 2.5
Row and column categories can also be included in the matrix in the following way:
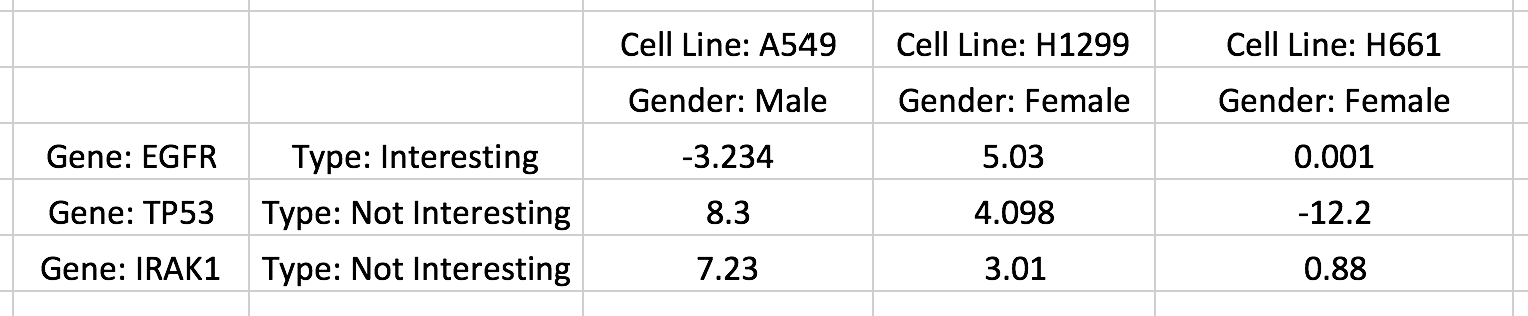
This screenshot of an Excel spreadsheet shows a single row category being added as an additional column of strings (e.g. 'Type: Interesting') and a single column category being added as an additional row of strings (e.g. 'Gender: Male'). Up to 15 categories can be added in a similar manner. Titles for row/column names or categories can be added by prefixing each string with Title:(note a space is required after the colon). For example the title of the column names is Cell Line and the title of the row categories is Gender .
Alternatively, row/column names and categories can be stored as Python tuples as shown below (or see tuple_cats.txt).
('Cell Line: A549', 'Gender: Male') ('Cell Line: H1299', 'Gender: Female') ('Cell Line: H661', 'Gender: Female')
('Gene: EGFR','Type: Interesting') -3.234 5.03 0.001
('Gene: TP53','Type: Not Interesting') 8.3 4.098 -12.2
('Gene: IRAK','Type: Not Interesting') 7.23 3.01 0.88
This format is easier to export from a Python Pandas DataFrame (see net.write_matrix_to_tsv). Note that 'titles' have been added to row and column names as well as row and column categories.
Several example tab-separated matrix files can be found in the txt directory. See example workflow or make_clustergrammer.py for examples of how to use Clustergrammer.py to generate a visualization json from these matrix files.
Optional clustergrammer.js Arguments
These arguments can also be passsed to Clustergrammer as part of the args object.
row_label and col_label: Pass strings that will be used as 'super-labels' for the rows and columns.
row_label_scale and col_label_scale: A number that will be used as a scaling factor that increases or decreases the size of row and column labels (as well as the font-size of the text).
super_label_scale: A number that will be used a a scaling factor that increases or decreases the size of the 'super-labels'.
ini_expand: Initialize the visualization in 'expanded' mode so that the sidebar controls are not visible.
opacity_scale: This defines the function that will map values in your matrix to opacities of cells in the visualization. The default is 'linear', but 'log' is also available.
input_domain: This defines the maximum (absolute) value from your input matrix that corresponds to an opacity of 1. The default is defined based on the maximum absolute value of any cell in your matrix. Lowering this value will increase the opacity of the overall visualization and effectively cutoff the visualization opacity at the value you choose.
do_zoom: This determines whether zooming will be available in the visualization. The default is set to true.
tile_colors: This determines the colors that indicate positive and negative values, respectively, in the visualization. The default are red and blue. The input for this is an array of hexcode or color names, e.g. ['#ED9124','#1C86EE'].
row_order and col_order: This sets the initial ordering of rows and columns. The default is clust. The options are
- alpha: ordering based on names of rows or columns
- clust: ordering based on clustering (covered here)
- rank: ordering based on the sum of the values in row/column
- rank_var: ordering based on the variance of the values in the row/column
ini_view: This defines the initial view of the clustergram. A clutergram can have many views available (discussed here) and these views generally consist of filtered versions of the clustergram.
sidebar_width: The width, in pixels, of the sidebar. The default is 150px.
sidebar_icons: This determines whether the sidebar will have icons for help, share, and screenshot. The default is true.
max_allow_fs: This sets the maximum allowed font-size. The default is set to 16px.
Clustergrammer was developed by Nicolas Fernandez at Icahn School of Medicine at Mount Sinai.